About Us
Group Website: https://ibots-bonn.de
iBOTS offers critical programming assistance to over 40 neuroscience labs across Bonn, Köln, Aachen, and Jülich in the iBehave network. Our mission is to enable data collection, processing, and analysis through swift training and knowledge dissemination. We focus on simplifying, automating, and disseminating data analysis pipelines using open-source software and programming environments.
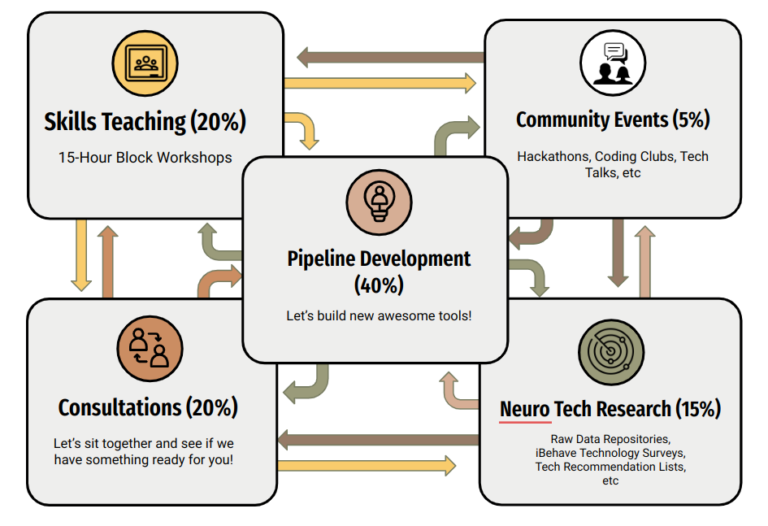
Unlike conventional engineering groups, iBOTS is a training and consulting platform that links researchers to open-source technologies and free resources. We’re committed to bridging the gap between scientific goals and technical execution by promoting knowledge exchange, collaborative development, and community building. This results in close relationships with researchers, custom, cost-effective neuroscience tech solutions, and high direct availability to scientists at all levels of their academic careers.
Resources
- Our Upcoming Workshops: https://ibots-bonn.de/teaching
- Reach out to us for Consulting! https://ibots-bonn.de/consulting/
- Use Our Teaching Materials for Self-Study or your own Computational Skills Teaching: https://learn.ibots-bonn.de/
Opportunities
Are you a Bachelor’s or Master’s student, and would like to learn research data management skills We are currently accepting application as part of the NeurotechEU Summer 2026 Program: https://www.neurotecheu.uni-bonn.de/en/studying/neurotecheu-internship-program/neurotecheu-internship-program





